2. Add a dataset¶
2.1. Quickstart: Use demo datasets¶
Download the demo datasets seen on the live EDB demo. Follow install instructions in the included README.
File: EDB_demodata.tar.gz (270Mb)
md5sum: d529107a0ac482f4d39f70ea0e7546aa
Note: If you installed EDB using the custom dockerfile, you already have the demo data.
2.2. Basic steps¶
Follow these steps to add a new dataset to the EDB. As an example, we use the demo BS-seq dataset from Lister, Mukamel et al. (2013). Science, which charts DNA methylation over human postnatal brain development.
Let us assume that EDB datasets are all located at <dataRoot>.
Step 1: dataset config: Create a master config file for the dataset.
Step 2: phenotype matrix: Add a table specifying sample phenotypes/metadata properties, and links to the sample-wise data
Step 3: data: Assemble the data files and any required index files (e.g. tbi for tabix files).
Step 4: group order: Add a file to specify the ordering of categorical variables
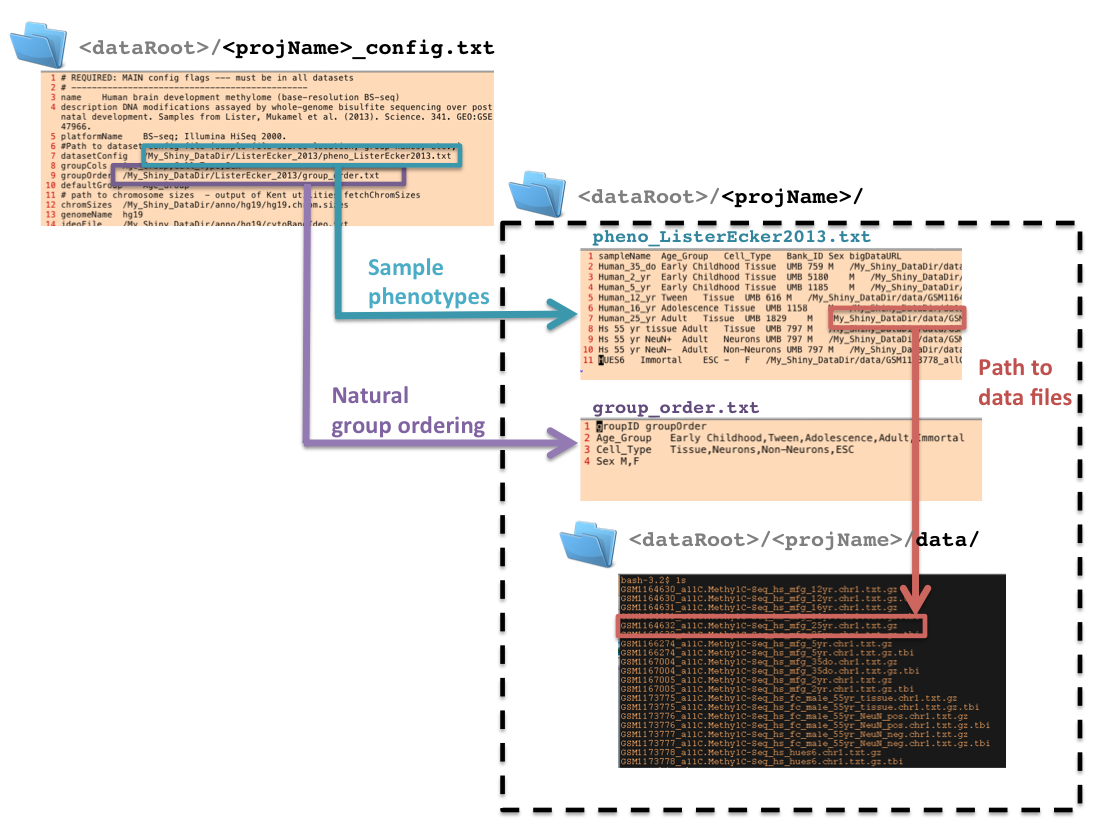
2.3. Example directory structure for datasets¶
This is what a simple project directory structure should look like when you are done. The example below assumes that all datasets and associated config files are located at <dataRoot>. Coloured arrows show how the master config file references the phenotype table and group order table.
In this example we have two datasets, one showing epigenetic dynamics in mouse brain development and the other, for human. Here, config_location.txt points to a directory which also contains all browser-related metadata and the data files themselves:
<config_location_points_here>
|
|---- mouseBrainDev_config.txt *symlink*
|---- humanBrainDNAMethylome_config.txt *symlink*
|---- mouseBrainDev/
|---- config.txt
|---- pheno.txt
|---- group_order.txt
|---- data
|---- GSM123456_mm9.bw
|---- GSM654321_mm9.bw
| ...
|---- GSM9999_mm9.bw
|---- humanBrainDNAMethylome/
|---- config.txt
|---- pheno.txt
|---- group_order.txt
|---- data
|----
|---- PCW4.bw
|---- midGestation_Cortex.bw
| ...
|---- PostPuberty_AnteriorCingulateCortex.bw
|----anno
2.4. Dataset config file¶
The dataset config file contains general metadata for the dataset. It is a tab-delimited file with two columns: a key (controlled word for config parameter) and value.
2.4.1. Format and required fields¶
| key | description |
|---|---|
name |
Dataset name; human-readable (~70 char) |
description |
Brief description of dataset; human-readable (~170 char) |
platformName |
Short description of platform on which data was generated; human-readable (~30 char) |
datasetConfig |
Path to sample phenotype table |
groupCols |
Column names of sample phenotype table <add-data-pheno> that EDB should allow samples to be grouped by; comma-separated list, case-sensitive |
groupOrder |
Path to group order file |
defaultGroup |
Name of group that serves as the default value in the Group by dropdown box of EDB interface; must be a column name of the sample phenotype table <add-data-pheno> , case-sensitive |
chromSizes |
Path to text file containing genome sequence sizes. This file should be the output of fetchChromSizes from Kent utilities at UCSC. |
genomeName |
Name of genome build ; must correspond to a UCSC release name <https://genome.ucsc.edu/FAQ/FAQreleases.html> |
ideoFile |
Path to text file corresponding to UCSC cytoBandIdeo table. Header? |
annoConfig |
Path to config file for genome annotation. TODO: Add link |
datatype |
Datatype. Currently, one of { bigwig , BSseq }. Indicates how data should be
processed; most single continuous traces (e.g. coverage, tiling microarrays) can use
the ‘bigwig’ setting. |
All the above fields are required and must not contain missing values.
2.4.2. Variables for different datatypes¶
bigwig:
This datatype has no required parameters.
BSseq:
BSseq__COV_POS: Column index of tabix file which contains base coverageBSseq__M_POS: Column index of tabix file which contains number of methylated reads at that base (“M” read count)
2.4.3. Example dataset config file¶
# REQUIRED: MAIN config flags --- must be in all datasets
# ----------------------------------------------
name Human brain development methylome (base-resolution BS-seq)
description DNA modifications assayed by whole-genome bisulfite sequencing over postnatal development. Samples from Lister, Mukamel et al. (2013). Science. 341. GEO:GSE47966.
platformName BS-seq; Illumina HiSeq 2000.
#Path to dataset config file (sample file source location, group names, etc.,)
datasetConfig /<dataRoot>/ListerEcker_2013/pheno_ListerEcker2013.txt
groupCols Age_Group,Cell_Type,Sex
groupOrder /<dataRoot>/ListerEcker_2013/group_order.txt
defaultGroup Age_Group
# path to chromosome sizes - output of Kent utilities fetchChromSizes
chromSizes /<dataRoot>/anno/hg19/hg19.chrom.sizes
genomeName hg19
ideoFile /<dataRoot>/anno/hg19/cytoBandIdeo.txt
annoConfig /<dataRoot>/anno/hg19/anno_config.txt
datatype BSseq
ylabel % methylation
#---------------------------------------------
# OPTIONAL: DATATYPE-specific; flags must have prefix <datatype>__
# Leave section blank if there are no flags.
#---------------------------------------------
BSseq__COV_POS 6
BSseq__M_POS 5
EDB requires that all dataset config files be located at the path specified in the special config_location.txt file. Recall that this file is at <EDBServerRoot>/config_location.txt.
2.5. Sample phenotype table¶
This tab-delimited file contains sample-wise metadata, including locations of data files. Each row should contain data for one sample, and each column should contain a unique type of metadata. Column order is unimportant to the browser. The browser expects the following columns, named exactly in this way:
2.5.1. Format¶
| column name | expected value |
|---|---|
sampleName |
unique identifier, no spaces |
bigDataURL |
absolute path to data source (e.g. .bw file) |
... |
all grouping columns as described in the grouping order file. |
2.5.2. Example phenotype table¶
sampleName Age_Group Cell_Type Bank_ID Sex bigDataURL
Human_35_do Early Childhood Tissue UMB 759 M /My_Shiny_DataDir/data/GSM116700
4_allC.MethylC-Seq_hs_mfg_35do.chr1.txt.gz
Human_2_yr Early Childhood Tissue UMB 5180 M /My_Shiny_DataDir/data/GSM1167005_allC.MethylC-Seq_hs_mfg_2yr.chr1.txt.gz
Human_5_yr Early Childhood Tissue UMB 1185 M /My_Shiny_DataDir/data/GSM1166274_allC.MethylC-Seq_hs_mfg_5yr.chr1.txt.gz
Human_12_yr Tween Tissue UMB 616 M /My_Shiny_DataDir/data/GSM1164630_allC.MethylC-Seq_hs_mfg_12yr.chr1.txt.gz
Human_16_yr Adolescence Tissue UMB 1158 M /My_Shiny_DataDir/data/GSM1164631_allC.MethylC-Seq_hs_mfg_16yr.chr1.txt.gz
Human_25_yr Adult Tissue UMB 1829 M /My_Shiny_DataDir/data/GSM1164632_allC.MethylC-Seq_hs_mfg_25yr.chr1.txt.gz
Hs 55 yr tissue Adult Tissue UMB 797 M /My_Shiny_DataDir/data/GSM1173775_allC.MethylC-Seq_hs_fc_male_55yr_tissue.chr1.txt.gz
Hs 55 yr NeuN+ Adult Neurons UMB 797 M /My_Shiny_DataDir/data/GSM1173776_allC.MethylC-Seq_hs_fc_male_55yr_NeuN_pos.chr1.txt.gz
Hs 55 yr NeuN- Adult Non-Neurons UMB 797 M /My_Shiny_DataDir/data/GSM1173777_allC.MethylC-Seq_hs_fc_male_55yr_NeuN_neg.chr1.txt.gz
HUES6 Immortal ESC - F /My_Shiny_DataDir/data/GSM1173778_allC.MethylC-Seq_hs_hues6.chr1.txt.gz
2.6. Groups and grouping order¶
The group_order.txt file is a tab-delimited file containing a table of two columns:
#. groupID: Group name, must match a column name in the phenotype matrix
#. groupOrder: Order in which group members must be shown. Comma-separated collection of values. All values for a given group must be specified here. The browser will return an error if any additional group members are found in the phenotype table but are not listed here.
In addition to these groups, the browser allows a non-grouping option - i.e. viewing sample-specific data - with “Grouping: (none)”.
2.6.1. Example group order file¶
groupID groupOrder
Tissue Brain,Sperm
Diagnosis Control,Schizophrenia,Bipolar disorder
TimeOfSampling Before_Treatment,During_Treatment,After_Treatment
For this dataset, the browser would show 4 grouping options: Tissue, Diagnosis, TimeOfSampling, (none).
The (none) option is automatically added, and allows samples to be inspected individually instead of being grouped.
3. Adding demo datasets¶
This section is added for completeness. Demo datasets have not yet been made publicly available. - SP 5 Sep 2014.
In this example, the demo data and config files are contained in a directory named “Shiny_BrowseR_Github_data”.
From the source machine, rsync the datasets to the machine where the EDB instance will be hsoted. The data directory should be in a location readable by the user shiny, which will run the EDB app.
rsync -avL --progress Shiny_BrowseR_Github_data $DEST_MACHINE:/.
On the destination machine, we update the path in the metadata files to reflect the path on our new machine. Replace the substitution sed command with one relevant to your source and destination paths.
Below we update the paths in the data dirs as well as the annotation directory (anno/hg19).
cd /Shiny_BrowseR_Github_data
rm *_config.txt
cd mTAG_BrainSperm
sed -i 's/\/src\/path\/dir/\/dest\/path/g' *.txt
cd ../ListerEcker_2013
sed -i 's/\/src\/path\/dir/\/dest\/path/g' *.txt
cd ../anno/hg19
sed -i 's/\/src\/path\/dir/\/dest\/path/g' anno_config.txt
Create a symlink to dataset-specific config files in the data directory
We change /srv/shiny-server/EDB/config_location.txt to point to our new data directory path: /Shiny_BrowseR_Github_data
At this point, refresh the EDB. If the “Choose dataset” dropdown box is populated, shiny can see the datasets.
If not, stop here and check the following:
- Is the data root directory in a location with read permissions for user “shiny”?
- Have the paths been correctly updated for all dataset directories?
- Is config_location.txt pointing to the correct data directory?
4. Adding custom annotation tracks¶
Annotation files are expected to live under <dataRootDir>/anno where <dataRootDir> is the directory to which config_location.txt points. Sources are organized by genome build as in the example below. EDB uses the BioConductor Gviz package to construct annotation objects.
Refresh the EDB browser page and reload dataset to see the listing of new annotation sources.
4.1. Directory structure for custom annotation¶
<dataRootDir>/
anno/
|------ hg19/
|------ cpgIslandExt.txt
|------ cytoBandIdeo.txt
|------ TxDb.Hsapiens.UCSC.hg19.refGene.sqlite
|------ anno_config.txt
|------ mm9/
|------ cpgIslandExt.txt
|------ LAD_NPC_mm9.txt
|------ anno_config.txt
4.2. anno_config.txt¶
anno_config.txt s used by EDB to get a listing of all available annotation for a genome build. EDB gets the genome build for the current dataset as the value of the genomeName variable in the dataset config file. It then refers to <dataRootDir>/<genomeName>/anno_config.txt for a list of all annotation available for that genome build. anno_config.txt is expected to be a tab-delimited file with rows representing each annotation source, and the following columns:
| column | description |
|---|---|
| trackName | one-word unique identifier for track |
| name | title of track as it would appear in EDB (<50 char) |
| description | (currently unused) |
| trackType | See allowed values below |
| defaultView | [dense|squish|full]. Similar to UCSC tracks. |
| bigDataURL | absolute path to source file |
| color | (currently unused) |
| format | See values in table below. |
| sizes | number between 0 and 1. determines the height of
the track. See plotTracks() method in Gviz |
Columns except description and color must not have missing values.
EDB currently supports the following file formats. Behaviour is undefined if the
format-trackType combinations below are not respected.
EDB format |
input file format / object | EDB trackType |
|---|---|---|
tabix |
tabix (.gz,.gz.tbi) | AnnotationTrack |
bigwig |
bigwig (.bw) | AnnotationTrack |
txdb |
BioC TranscriptDB object (.sqlite) | GeneRegionTrack |
Visit these pages to learn more about the tabix , bigwig and BioC TranscriptDB format/objects.
4.2.1. Sample anno_config.txt file¶
trackName name description trackType defaultView bigDataURL color format sizes
cpgIslands CpG Islands CpG Islands AnnotationTrack dense /home/docker/EDB_demodata/anno/hg19/cpgIslandExt.bed.gz green tabix 0.15
refGene RefSeq genes RefSeq genes GeneRegionTrack squish /home/docker/EDB_demodata/anno/hg19/TxDb.Hsapiens.UCSC.hg19.refGene.sqlite mediumblue TxDb 0.1